 |
|
The McMahan lab excavates the junction between nerve and muscle, seeking a microscopic go-between
By Rosanne Spector
Photographs Leslie Williamson
Some architecture lovers travel to Mexico to see the pyramids of Teotihuacan. Others visit Italy's Cathedral of Pisa, with its leaning tower. But neurobiology professor Jack McMahan, PhD, considers a computer screen in his lab at Stanford to be the best place to view great architecture.
That's where McMahan can ogle the structures that intrigue him most: the beams and struts that form a microscopic architecture within our cells and tissues.
On the screen he has an unsurpassed view of the inner organization of a cell's structure and the tissue components that link one cell to another – a view made possible by an imaging method McMahan and his team have been perfecting for most of the past decade.
Unlike other imaging techniques, theirs is particularly suited to reveal the shapes and relationships of the groups of proteins, known as macromolecules, that direct most of our bodies' activities. As McMahan hoped to find when he began developing the technology, the architecture is marvelous.
"It seems that in all of our cells, from the big toe to the top of the head, there is a continuous scaffold of macromolecules on which much of the chemistry that keeps us alive is carried out," says McMahan.
McMahan has a favorite destination in our molecular terrain.For the past 32 years, he has been investigating the meeting place between nerve and muscle, the neuromuscular synapse.
During this time he and his associates discovered and characterized a protein so important it has spawned dozens of full-fledged research projects around the world. That protein, agrin, leaps from the end of a nerve cell to the structure next door – a muscle fiber. Once there it spurs the muscle fiber to make and maintain the cellular machinery that responds to the neurotransmitter acetylcholine.
Thanks to this machinery – constructed of many macromolecules and organelles – our muscles contract when zapped by acetylcholine molecules sent by a neighboring nerve ending.
What's the big deal about agrin? McMahan explains: "Without agrin we couldn't move. In fact, we couldn't even get our chest muscles to make us breathe." The cellular machinery agrin creates is that important.
While many researchers continue to elucidate the biochemical properties of agrin, McMahan has moved on. He wants to spy on agrin and its macromolecular associates in their natural habitat.
If McMahan and his team perfect their imaging technique, they won't be the only ones to benefit. Because while other techniques such as crystallography and NMR can reveal details of molecules in isolation, no method other than the one McMahan's team is developing allows researchers to zoom in on such tiny structures "on location" as it were.
Harvard neuroanatomy professor emeritus Sanford L. Palay, MD, puts it this way: "Jack and his group have a very strong curiosity about what goes on at the tiny junction between the nervous system and the muscle. That's driving them to develop a method for reconstructing the structures there. It's very hard work to do, but they've got the method down for the synapse. Eventually that same method might be used by others to reconstruct such structures throughout an organism."
Constructive musing: The basic methodology used by McMahan and his team works something like a CT scan, but instead of using X-ray images to build a 3-D reconstruction it uses electron micrographs. Before McMahan began, others used the technique for characterizing the structure of macromolecules after they had been extracted from tissues and then purified.
The seed of the idea to use the technology for the more complicated task of viewing macromolecules in their native tissues was a scenario he came across in a thriller – Michael Crichton's "Rising Sun" – in which investigators identify a murderer using video forensics. They reconstruct a man's face from murky reflections caught on a surveillance video.
McMahan lab microscopist Bob Marshall remembers McMahan musing about the technique's possibilities. Says Marshall: "Jack asked if it would be possible for us to do something similar with electron microscopy for structures in tissue sections too small to be seen clearly by the usual methods. I said I thought we could do it, and off we went."
So in 1993 McMahan mustered a research group that came to consist of research associate Marshall, graduate student Mark Harlow (now a postdoc in the lab) and two postdocs: David Ress, PhD (now a senior staff scientist in the labs of both McMahan and associate professor of psychology David Heeger, PhD) and Arne Stoschek, PhD (now senior director of the Electronics Research Lab at Volkswagen of America in Sunnyvale, Calif.). Together they joined the group of just a dozen or so research labs worldwide striving to develop the new technology of electron microscope tomography.
The technique's advantage over conventional electron microscopy of tissue sections is that it provides three-dimensional information. And the benefit of McMahan's 3-D strategy over others is that it shows how macromolecules interact with their neighbors – be they cell membranes, organelles or other macromolecules – within the tissue they normally inhabit.
McMahan's approach uses a series of two-dimensional images of a section made at various tilt angles to reconstruct the three-dimensional volume. Because the researchers are pushing the microscope to its limits, obtaining a single usable set of 140-180 images can take a full day. The microscope must work perfectly or the effort's a bust.
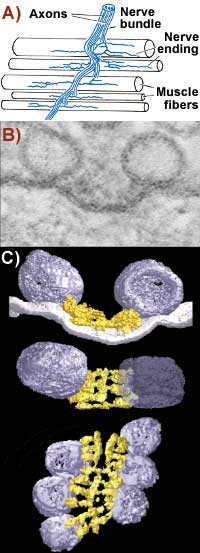 |
A) The neuromuscular junction: McMahan studies the action where nerve endings meet muscle. B) Active zone material at the neuromuscular junction, imaged using conventional electron microscopy. Bar is 50 nanometers. C) McMahan's molecular CT scanning reveals more than microscopy. Tilting the image reveals ribs and beams in the active zone and shows how they interact with the vesicles. |
|
Software helps but brains still needed: Once they get a good 3-D reconstruction, the team members use software Ress and Stoschek developed to analyze the arrangement of the
macromolecules, membranes and organelles. They begin searching for arrangements predicted from biochemical or electrophysiological methods. If their efforts fail to confirm these predictions, "then the fun begins," says McMahan.
At that point the researchers begin developing testable hypotheses as to what the new results mean in terms of how the neuromuscular synapse works, says McMahan. The researchers log hour after hour in front of a computer screen using the software to travel back and forth through the reconstructed tissue section trying to recognize the organizational plan.
"This takes time and this takes concentration," says McMahan. "The macromolecular scaffold in a tissue section from a neuromuscular synapse is imposing in its complexity. Some components are tightly intertwined like vines; others are globular, like basketballs; and others are loosely meshed like chicken wire. But common sense dictates they must be highly organized relative to one another.
"You'll sit there for hours, sometimes days, trying to pick up the slightest trace of organization and then you'll work even longer fully exposing it."
Luckily, McMahan loves the work and his talented team is as dedicated as he is. Ress and Stoschek worked out the computer algorithms; Marshall and McMahan prepare the tissue and collect the images; and Harlow uses the algorithms to trace the macromolecular arrangements within the reconstructions.
"The result of our shared effort has been aesthetically as well as intellectually exhilarating. Biological structure at nanometer scale is astoundingly beautiful," says McMahan.
 |
|
Jack McMahan, PhD, at an electron microscope. |
|
Mere blobs no more: The team's efforts have paid off with structural details about the portion of the macromolecular scaffold, called active zone material, massed in the cytoplasm of the nerve ending just across the gap between the ending and the muscle fiber. The best that conventional electron microscopy could do was portray the stuff as small blobs.
Now a much clearer picture emerges. By concentrating on a specific region in each clump of active zone material McMahan's team has used its technique to reveal that the material consists of an ordered array of distinct components they have named "ribs," "beams" and "pegs." Each is about 5 nanometers in diameter and about 10 times as long. The organization and relationships of ribs, beams and pegs together with predictions as to the identities of some of the proteins they contain helps explain how the nerve impulse causes release of acetylcholine from nerve endings to stimulate the muscle fibers. The group detailed its findings in an article published in Nature last year (Jan. 25, 2001).
These days, the McMahan lab is testing its predictions as to the function of these macromolecules and the identity of the proteins in them. One strategy: bathe tissue samples in botulinum toxins. Biochemistry has shown that the toxins degrade certain proteins involved in the release of acetylcholine from nerve endings, leading to paralysis in living tissue. Since the researchers propose that the ribs contain such proteins, they'll be interested in seeing whether botulinum disintegrates the ribs.
Meanwhile they'll fine-tune their methods for making reconstructions and analyzing data. Their long-term goal is to characterize the entire 3-D architecture of the neuromuscular synapse at macromolecular resolution and then to map onto that framework the positions of proteins such as agrin. One way to do that may be to apply mapping techniques that have been successfully used with 2-D electron microscopy. But McMahan suspects that more precise mapping methods will have to be developed to take full advantage of the increased spatial resolution provided by the 3-D reconstructions.
"We have started trying to work out a way to study the material in the synaptic cleft. It's like a three-dimensional cobweb spun by a master spider. It's just as beautiful, but it's much more complicated than anything we have previously encountered," McMahan says.
He says it could take 10 years or more until he and his team catch agrin moving across the 50-nanometer-wide gap from nerve to muscle and capture agrin's macromolecular associations at each step along the way.
But he won't mind spending hours at the computer monitor. He expects he will enjoy the sights.
Comments? Contact Stanford Medicine at

